Đào tạo & nghiên cứu
Developing Multiplex Real-Time PCR for the Identification of Circulating Streptococcus pneumoniae serotypes
H.T.Pham[1]*, T.K.T.Le[1,2], D.K.Tran[1,2], T.H.T.Nguyen[1,2], Q.D.Ha[1,2], A.H.Pham[1,2], V.H.Pham[1,3]
[1] Vietnam Research and Development Institute of Clinical Microbiolgy, Vietnam
[2] Nam Khoa Biotek Co. Ltd., Vietnam
[3] Phan Chau Trinh University, Vietnam
*First author
Background
More than 90 serotypes of S. pneumoniae have been reported until now. Determining these serotypes is crucial for monitoring antibiotic resistance, assessing vaccine coverage, and identifying the prevalent serotypes associated with diseases. However, implementing traditional methods as the Quelling reaction is challenging due to the requirements of the specific antisera for the different serotypes. To address this issue, the WHO proposes a solution with Multiplex PCR (MPL-PCR), targeting specific sequences on specific genes at the capsular synthesis locus. Many researchers suggest Multiplex real-time PCR (MLP rPCR) as a simpler technique without electrophoresis. Currently, no research has developed MPL-rPCR to detect all serotypes, as in the WHO’s MPL-PCR method.
| Aim of the study
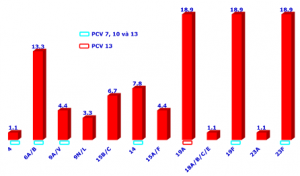
We aim to develop the MPL-rPCR to detect all serotypes as the WHO suggests by designing specific primers and probes targeting individual serotypes. Materials and methods We designed the specific primers and probes targeting the specific sequences at the capsular synthesis locus and from that, developed a 10-multiplex assay, each detects 4 serotypes or serogroups. This MLP rPCR can be performed on the S. pneumonia2 isolates, as well as on various samples, including DNA extractions from blood, cerebrospinal fluid (CSF) or sputum. We subsequently analyzed the results and compared to the PCR method suggested by the WHO. ResultsWe gathered 90 S. pneumoniae isolates from blood and CFS, along with 63 sputum samples. From the blood and CFS isolates, with our MPL-rPCR, we identified 12 serotypes or serogroups. The most prevalent were 19A, 19F, and 23F, each accounting for 18.9%, followed by 6A/B at 13.3%. Other serotypes were each detected in less than 10%, including 14 (7.8%), 15B/C (6.7%), 15A/F (4.4%), 9A/V (4.4%), 9N/L 3.3%), 4 (1.1%), 18 A/B/C/E (1.1%), và 23A (1.1%). In the 63 sputum samples collected from inpatients with pneumonia, 76.2% of the samples were detected the pneumococcal serotypes that were belonged to 16 serotypes or serogroups. In which the serotype 9N/L was dominated (23.8%), followed by 24 A/F at 7.9%, and 39, 10F/C/33C, each at 6.3%. Our findings were comparable with the results obtained from the PCR recommended by the WHO and the agreement that we had received was 100%. ConclusionsThe MLP rPCR technique that we develop is proficient in detecting S. pneumoniae serotypes, aligning with WHO recommendations. If widely and routinely applied, this technique facilitates addressing antibiotic resistance, vaccine coverage, and advancing research on serotypes associated with various clinical conditions Keywords: Pneumococcal serotypes, Multiplex Real-time PCR
|
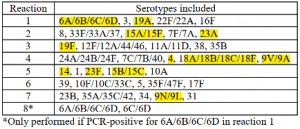
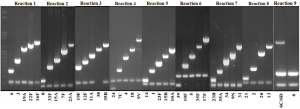
Table 1: PCR-serotyping scheme for USA based on current serotype prevalence (recommended by WHO), and the S. pneumoniae serotype must be identified by agarose electrophoresis
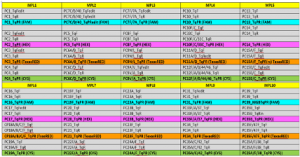
Table 2: The MPL-rPCR for identification of the S. pneumoniae serotype is based on 10 MPL-rPCR mix
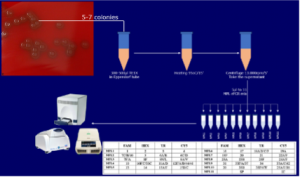
Figure 1: The MPL-rPCR procedure for identification of the S. pneumoniae serotype from the bacterial colonies
Graph 1: The serotypes of S. pneumoniae causing meningitis and septicemia in children (N=90) identified by the MPL-RPCR
|
REFERENCES
- World Health Organization & Centers for Disease Control and Prevention (S.). (2011). Laboratory methods for the diagnosis of meningitis caused by neisseria meningitidis, streptococcus pneumoniae, and haemophilus influenzae: WHO manual, 2nd ed. World HealthOrganization. https://iris.who.int/handle/10665/70765
- Pimenta FC, Roundtree A, Soysal A, Bakir M, du Plessis M, Wolter N, von Gottberg A, McGee L, Carvalho Mda G, Beall B. Sequential triplex real-time PCR assay for detecting 21 pneumococcal capsular serotypes that account for a high global disease burden. J Clin Microbiol. 2013 Feb;51(2):647-52. doi: 10.1128/JCM.02927-12. Epub 2012 Dec 5. PMID: 23224094; PMCID: PMC3553924.
- Tarrago , A. Fenoll, D. Sa´nchez-Tatay, L. A. Arroyo, C. Mun˜oz-Almagro, C. Esteva, W. P. Hausdorff, J. Casal1 and I. Obando. Identification of pneumococcal serotypes from culture-negative clinical specimens by novel real-time. Clinical Microbiology and Infection. Volume 14, Issue 9, September 2008, Pages 828-834. https://doi.org/10.1111/j.1469-0691.2008.02028.x